Often in Natural Language Processing (NLP) in Healthcare and other sophisticated fields, we often judge a model’s success based on how accurate it is. But this approach can be misleading because it doesn’t show how well the model handles real-world language. To truly know if a model is good, we need to test its robustness — how well it deals with different kinds of changes in the text it’s given. This is where Robustness Testing comes in. It’s like giving the model challenges to see if it can handle them. Imagine a ship sailing through rough waters — that’s the kind of test we’re talking about for NLP models. In this blog, we’ll dive into LangTest, a way to go beyond just accuracy and explore how well Named Entity Recognition models can handle the twists and turns of real language out there.
In addition, the article delves into the implementation details of the med7 and ner posology models and demonstrates their usage. We showcase the evaluation of model performance using LangTest’s features, conducting tests on both models for Robustness and Accuracy. Finally, we compare the performance of these models, providing valuable insights into their strengths and weaknesses in healthcare NER tasks.
What is Named Entity Recognition?
Named Entity Recognition (NER) is a natural language processing (NLP) technique that involves identifying and classifying named entities within text. Named entities are specific pieces of information such as names of people, places, organizations, dates, times, quantities, monetary values, percentages, and more. NER plays a crucial role in understanding and extracting meaningful information from unstructured text data.
The primary goal of Named Entity Recognition is to locate and label these named entities in a given text. By categorizing them into predefined classes, NER algorithms make it possible to extract structured information from text, turning it into a more usable format for various applications like information retrieval, text mining, question answering, sentiment analysis, and more.

Why Robustness Testing Matters?
Consider the scenario of Named Entity Recognition (NER). It involves examining how the model identifies entities within text documents.
For instance, we can investigate how the model performs on a sentences, and then deliberately transform the text into all uppercase and lowercase and test the model again on.
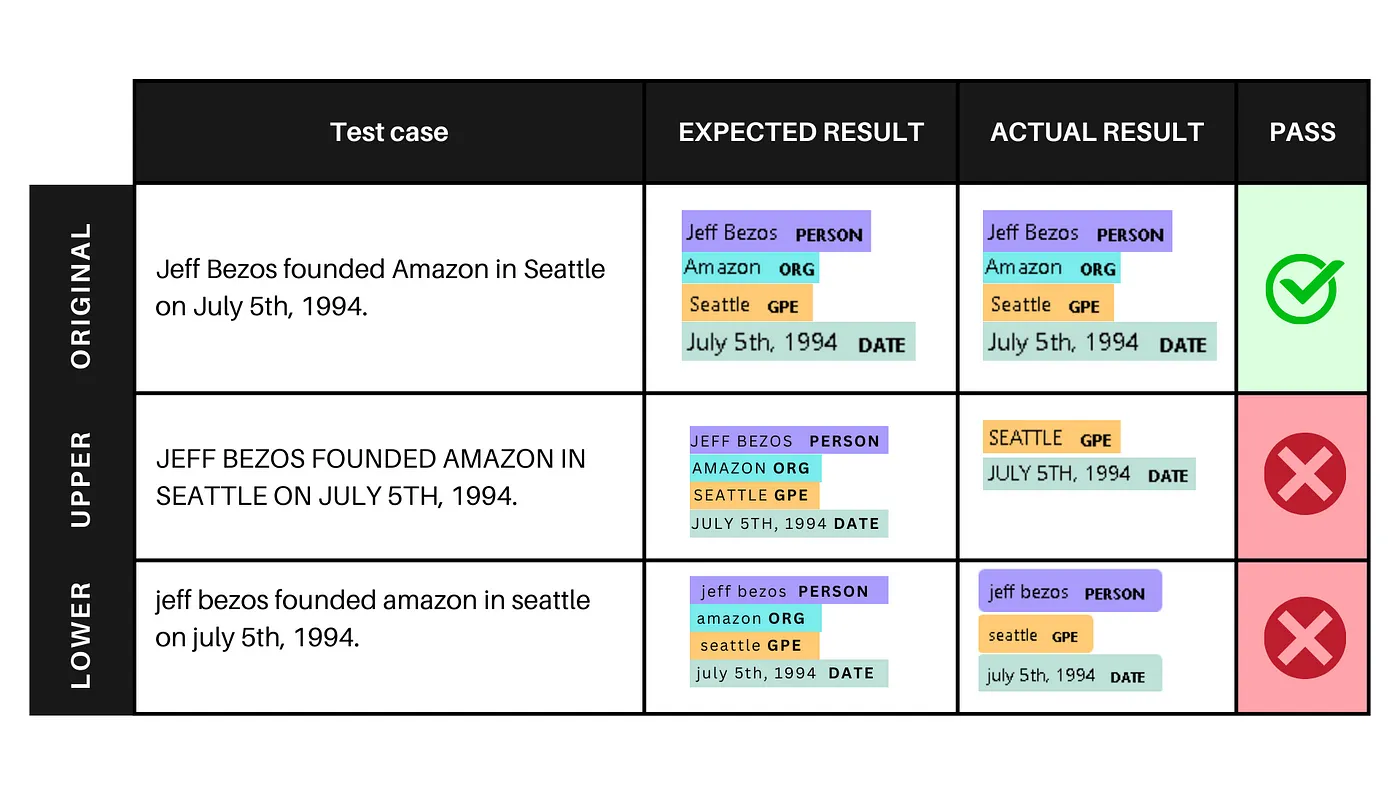
The output displays the identified named entities in each version of the sentence. Here’s what the output means for each example:
- Example 1: The model accurately identifies named entities in the original sentence, where entity types and case are consistent.
- Example 2: When the sentence is in all uppercase, the model retains some entity recognition but fails to identify “JEFF BEZOS” as a person and “AMAZON” as an organization due to the altered case.
- Example 3: When the sentence is in all lowercase, the model retains some entity recognition but fails to identify “amazon” as an organization due to the altered case.
This allows us to observe any variations in the model’s prediction quality. By conducting similar experiments with various perturbations, such as introducing typos or altering the writing style, we gain insights into the model’s robustness and its ability to generalize beyond the training data.
Robustness testing evaluates a model’s ability to handle variations and uncertainties commonly found in real-world data. It’s an essential step in ensuring that NER models, and machine learning models in general, are reliable, accurate, and capable of performing well across different situations.
Beyond accuracy, it’s imperative to examine a model’s behavior under different perturbations and real-world conditions. This holistic evaluation enables us to gauge a model’s adaptability and resilience in the face of challenges that may not be adequately captured by accuracy metrics alone. By including such robustness testing in our evaluation pipeline, we ensure that our models are not only accurate in controlled settings but also reliable and capable of performing well across diverse scenarios. Therefore, apart from checking accuracy, prioritizing the assessment of a model’s robustness is a crucial step towards building trustworthy and effective NER models.
LangTest
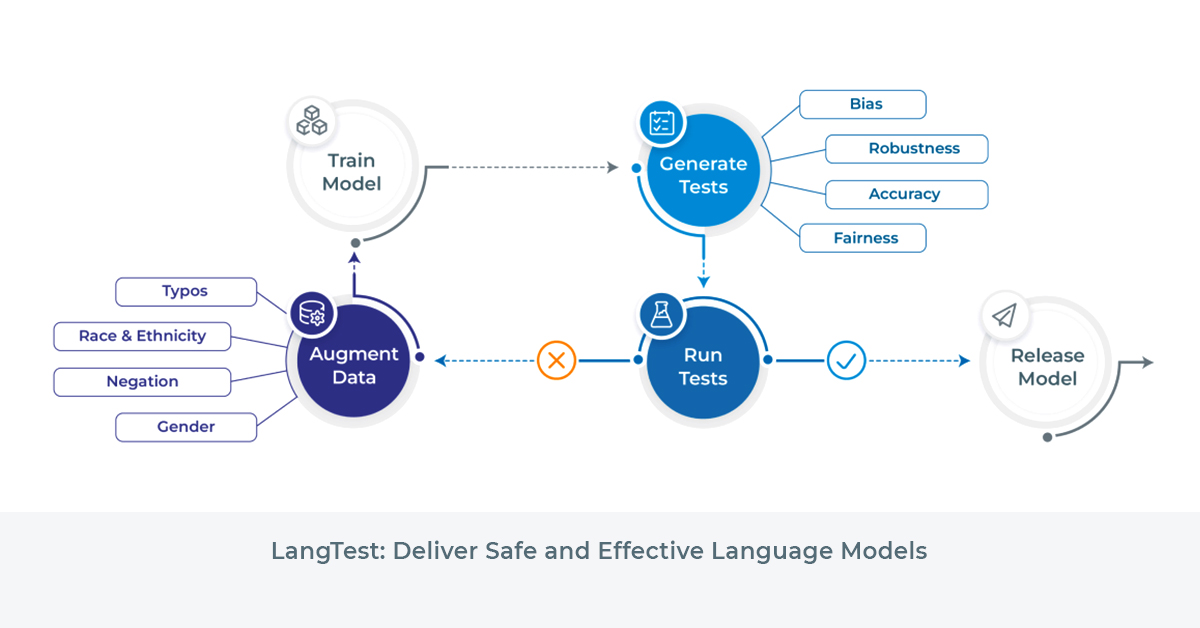
LangTest is an open-source Python library that facilitates the evaluation of NLP models. It provides a comprehensive set of tests for NER, text classification, question-answering, and summarization models. With over 50 out-of-the-box tests, LangTest covers various aspects such as robustness, accuracy, bias, representation, and fairness.
You can find the LangTest library on GitHub, where you can explore its features, documentation, and the latest updates. Additionally, for more information about LangTest and its capabilities, you can visit the official website at langtest.org.
Now we are going to take two healthcare NER models such as ner posology and med7, we can gain insights into their performance and make informed decisions about their suitability for specific use cases.
Model Overview
Named Entity Recognition (NER) models play a crucial role in extracting relevant information from clinical text, enabling tasks such as medication extraction and dosage identification in healthcare.The two NER models we will examine are ner posology and med7, where both the models recognizes seven categories, including Drug, Duration, Strength, Form, Frequency, Dosage, and Route.
ner posology — Spark NLP Models Hub
The ner posology model by John Snow Labs is specifically designed for posology NER tasks in the healthcare domain. It utilizes the embeddings_clinical word embeddings model. This pretrained deep learning model demonstrates strong performance in extracting medication-related information from clinical text.
med7 — GitHub Repository
med7 is a powerful clinical NER model that offers a comprehensive solution for extracting relevant information from healthcare text. Trained on the MIMIC-III dataset, it is compatible with spaCy v3+ . The model’s details can be found in the paper titled “Med7: a transferable clinical natural language processing model for electronic health records” authored by Andrey Kormilitzin, Nemanja Vaci, Qiang Liu, and Alejo Nevado-Holgado, published as an arXiv preprint in 2020.
Implementation: Using the ner posology Model
Ensure you have acquired and configured the required license keys for Spark NLP’s . Then, create Spark NLP and Spark Session using the official documentation as your guide.
To process the text and extract the desired entities using the ner posology model, you need to build an NLP pipeline.
You can find a comprehensive guide on how to build this pipeline by clicking here.
ner_posology_langtest = nlp_pipeline.fit(spark.createDataFrame([[""]]).toDF("text"))
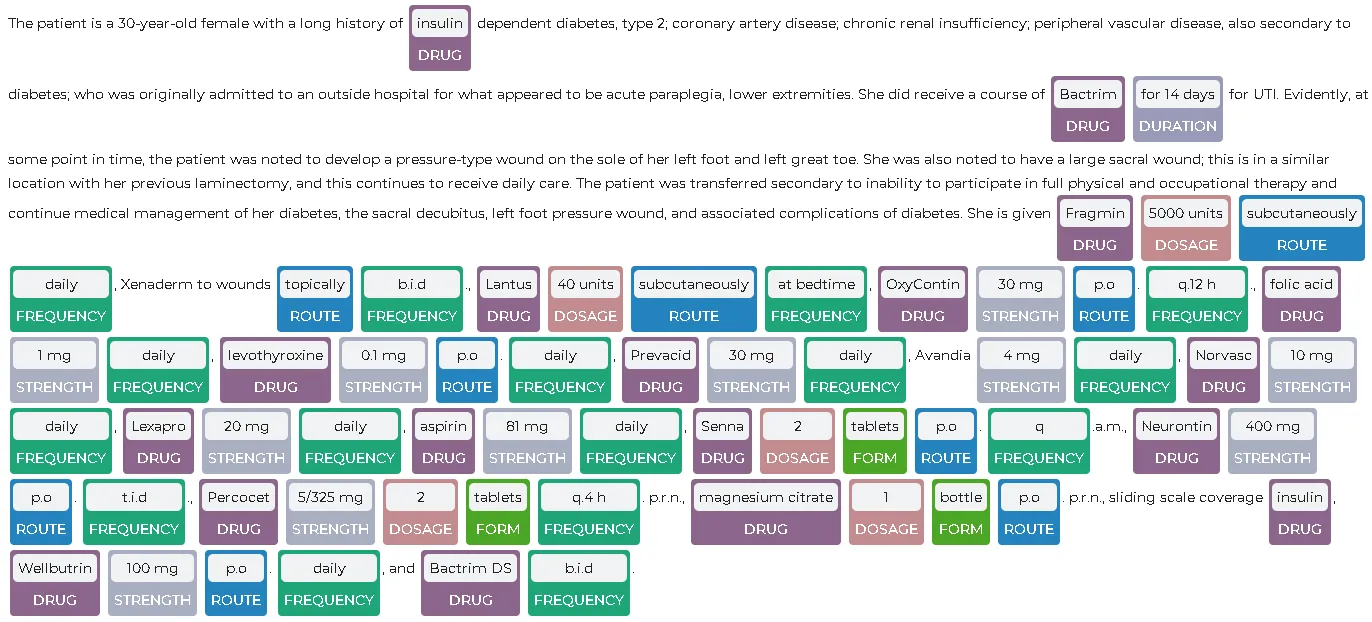
result = model.transform(spark.createDataFrame([['The patient is a 30-year-old female with a long history of insulin dependent diabetes, type 2; coronary artery disease; chronic renal insufficiency; peripheral vascular disease, also secondary to diabetes; who was originally admitted to an outside hospital for what appeared to be acute paraplegia, lower extremities. She did receive a course of Bactrim for 14 days for UTI. Evidently, at some point in time, the patient was noted to develop a pressure-type wound on the sole of her left foot and left great toe. She was also noted to have a large sacral wound; this is in a similar location with her previous laminectomy, and this continues to receive daily care. The patient was transferred secondary to inability to participate in full physical and occupational therapy and continue medical management of her diabetes, the sacral decubitus, left foot pressure wound, and associated complications of diabetes. She is given Fragmin 5000 units subcutaneously daily, Xenaderm to wounds topically b.i.d., Lantus 40 units subcutaneously at bedtime, OxyContin 30 mg p.o. q.12 h., folic acid 1 mg daily, levothyroxine 0.1 mg p.o. daily, Prevacid 30 mg daily, Avandia 4 mg daily, Norvasc 10 mg daily, Lexapro 20 mg daily, aspirin 81 mg daily, Senna 2 tablets p.o. q.a.m., Neurontin 400 mg p.o. t.i.d., Percocet 5/325 mg 2 tablets q.4 h. p.r.n., magnesium citrate 1 bottle p.o. p.r.n., sliding scale coverage insulin, Wellbutrin 100 mg p.o. daily, and Bactrim DS b.i.d.']], ["text"]))To visualize the extracted entities, you can use the NerVisualizer class from the sparknlp_display library. you can refer to the Spark NLP Display section in the documentation.
Implementation: Using the med7 Model
Install the model:
!pip install https://huggingface.co/kormilitzin/en_core_med7_lg/resolve/main/en_core_med7_lg-any-py3-none-any.whl
To utilize the med7 model, you can follow these steps:
import spacy
med7 = spacy.load("en_core_med7_lg")
col_dict = {}
seven_colours = ['#e6194B', '#3cb44b', '#ffe119', '#ffd8b1', '#f58231', '#f032e6', '#42d4f4']
for label, colour in zip(med7.pipe_labels['ner'], seven_colours):
col_dict[label] = colour
options = {'ents': med7.pipe_labels['ner'], 'colors':col_dict}
text = 'The patient is a 30-year-old female with a long history of insulin dependent diabetes, type 2; coronary artery disease; chronic renal insufficiency; peripheral vascular disease, also secondary to diabetes; who was originally admitted to an outside hospital for what appeared to be acute paraplegia, lower extremities. She did receive a course of Bactrim for 14 days for UTI. Evidently, at some point in time, the patient was noted to develop a pressure-type wound on the sole of her left foot and left great toe. She was also noted to have a large sacral wound; this is in a similar location with her previous laminectomy, and this continues to receive daily care. The patient was transferred secondary to inability to participate in full physical and occupational therapy and continue medical management of her diabetes, the sacral decubitus, left foot pressure wound, and associated complications of diabetes. She is given Fragmin 5000 units subcutaneously daily, Xenaderm to wounds topically b.i.d., Lantus 40 units subcutaneously at bedtime, OxyContin 30 mg p.o. q.12 h., folic acid 1 mg daily, levothyroxine 0.1 mg p.o. daily, Prevacid 30 mg daily, Avandia 4 mg daily, Norvasc 10 mg daily, Lexapro 20 mg daily, aspirin 81 mg daily, Senna 2 tablets p.o. q.a.m., Neurontin 400 mg p.o. t.i.d., Percocet 5/325 mg 2 tablets q.4 h. p.r.n., magnesium citrate 1 bottle p.o. p.r.n., sliding scale coverage insulin, Wellbutrin 100 mg p.o. daily, and Bactrim DS b.i.d.'
doc = med7(text)
spacy.displacy.render(doc, style='ent', jupyter=True, options=options)
[(ent.text, ent.label_) for ent in doc.ents]
By conducting a series of tests using the LangTest Python library, we can evaluate the Robustness and Accuracy of the ner posology and med7 models. These tests will help us understand their performance and limitations in healthcare applications.
Evaluation Metrics
To thoroughly assess the performance and compare the Robustness and Accuracy of the ner posology and med7 models, we will employ the LangTest Python library. LangTest provides a wide range of tests for evaluating NER models, including Robustness and Accuracy.
Robustness Testing
Robustness testing aims to evaluate the ability of a model to maintain consistent performance when faced with various perturbations or modifications in the input data. For NER models, this involves understanding how changes in capitalization, punctuation, typos, contractions, and contextual information affect their prediction performance.
Accuracy Testing
Accuracy testing is crucial for evaluating the overall performance of NER models. It involves measuring how well the models can correctly predict outcomes on a test dataset they have not seen before. LangTest provides various accuracy tests to evaluate precision, recall, F1 score, micro F1 score, macro F1 score, and weighted F1 score.
To learn more about each test and its implementation, you can refer to the tutorials available in the LangTest website.All code and results displayed in this blogpost is available to reproduce right here.
Evaluating Model Performance with LangTest
Now that we have introduced the two prominent NER healthcare models, ner posology and med7, it is essential to evaluate and test their performance. Evaluating these models allows us to understand their strengths, weaknesses, and overall suitability for extracting relevant information from clinical text. To accomplish this, we will utilize the LangTest.
To use LangTest, you can install it using pip
# Installing LangTest version 1.3.0 !pip install langtest==1.3.0
First we will import the Harness class from the LangTest library in the following way.
from langtest import Harness
Harness class from within the module, that is designed to provide a blueprint or framework for conducting NLP testing, and that instances of the Harness class can be customized or configured for different testing scenarios or environments.
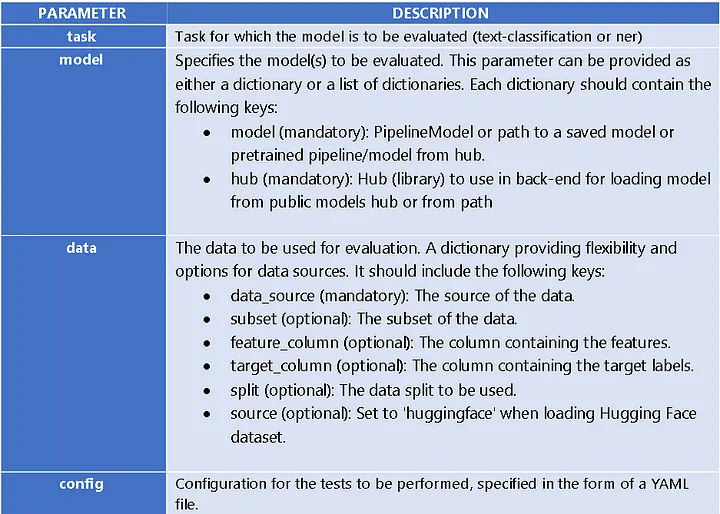
Here is a list of the different parameters that can be passed to the Harness class:
Testing ner posology model
Instantiate the Harness Class
we have instantiated the Harness class to perform NER testing on the ner posology model. We have specified the test data, set the task to “ner”, and provided the model.
harness = Harness(
task = "ner",
data={"data_source":"sample-test.conll"},
model={"model":ner_posology_langtest,"hub":"johnsnowlabs"}
)Configure the Tests
We can use the .configure() method to manually configure the tests we want to perform.
Here we are going to configure the harness to perform Robustness and Accuracy tests. For each test category, we will specify the minimum pass rates and additional parameters where applicable.
# Define the test configuration function
def configure_tests():
robustness_tests = [
'uppercase', 'lowercase', 'titlecase', 'add_punctuation', 'strip_punctuation',
'add_slangs', 'dyslexia_word_swap', 'add_abbreviation', 'add_speech_to_text_typo',
'number_to_word', 'add_ocr_typo', 'adjective_synonym_swap'
]
accuracy_tests = [
'min_precision_score', 'min_recall_score', 'min_f1_score', 'min_micro_f1_score'
]
min_pass_rate = 0.70
min_score = 0.70
tests_configuration = {
'defaults': {'min_pass_rate': 0.70},
'robustness': {test: {'min_pass_rate': min_pass_rate} for test in robustness_tests},
'accuracy': {test: {'min_score': min_score} for test in accuracy_tests}
}
harness.configure({
'tests': tests_configuration
})Test Case Generation and Execution
#Configure the Tests configure_tests() # Call the test configuration function # Generating the test cases harness.generate() # This method automatically generates test cases based on the provided configuration. # Running the tests harness.run() # This method is called after harness.generate() and is used to run all the tests. It returns a pass/fail flag for each test. # Generated Results harness.generated_results() # To obtain the generated results.
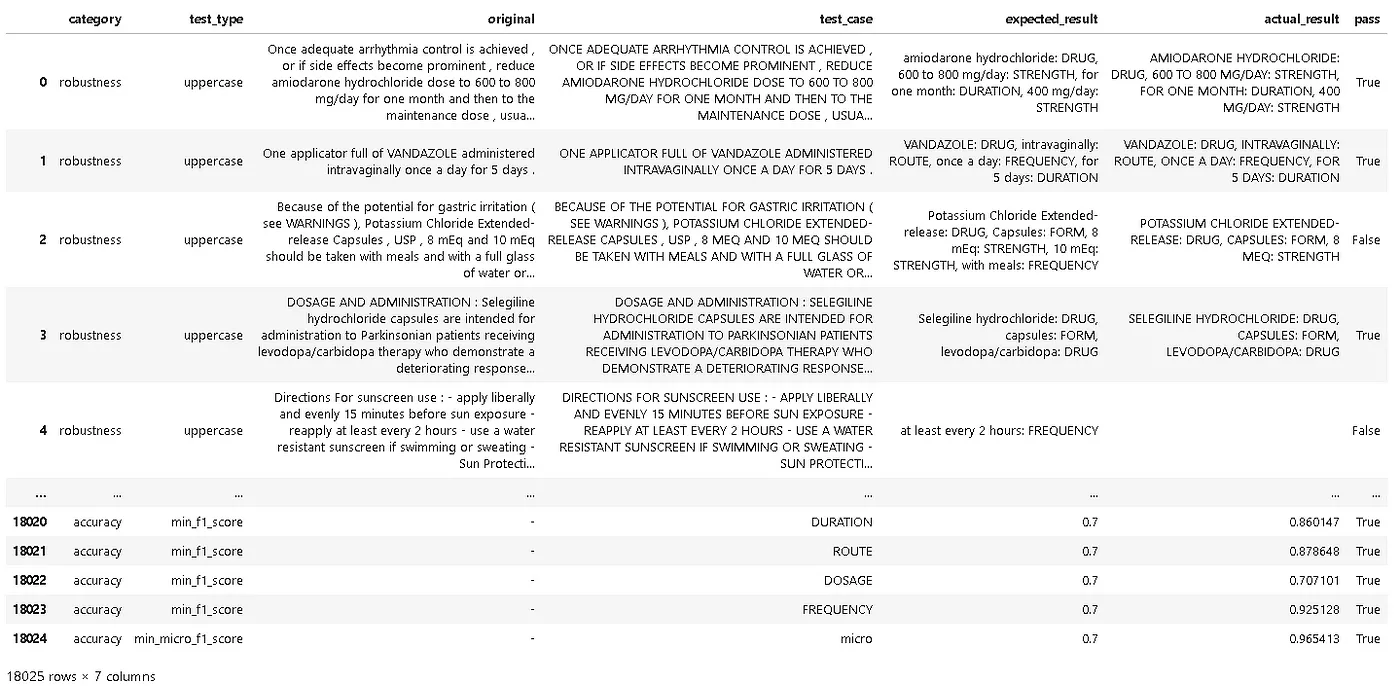
Report of the tests
harness.report()
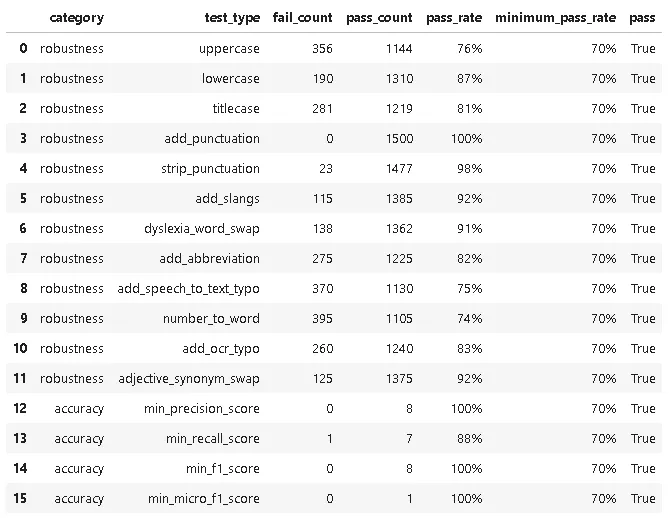
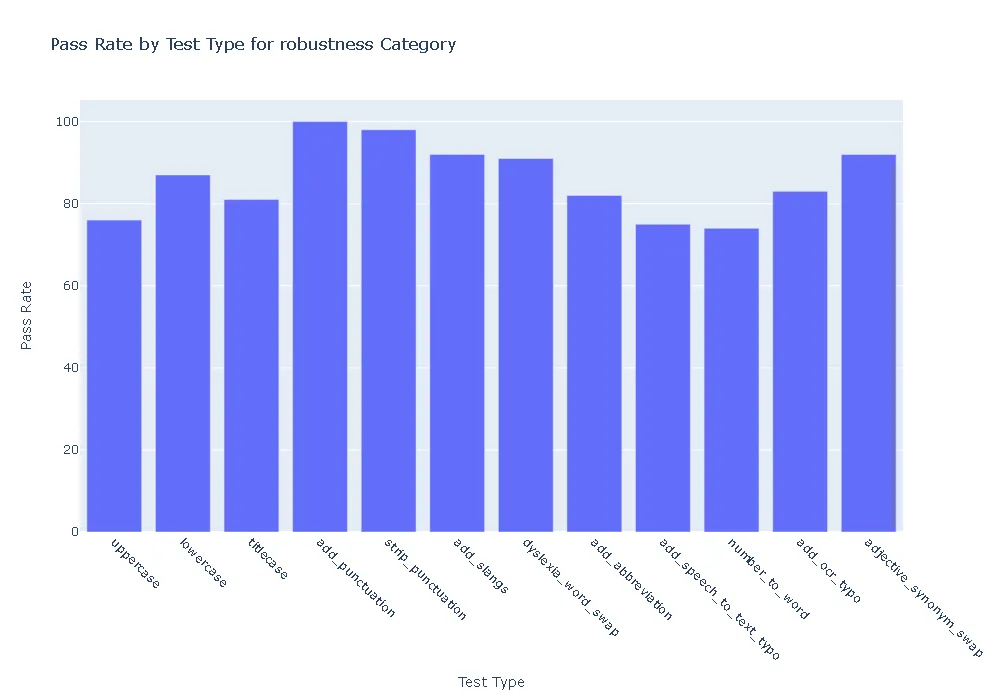
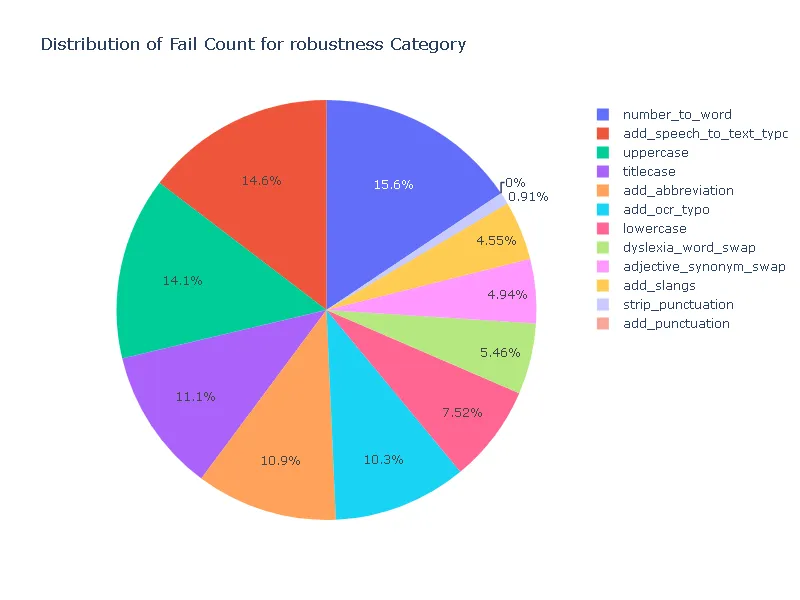
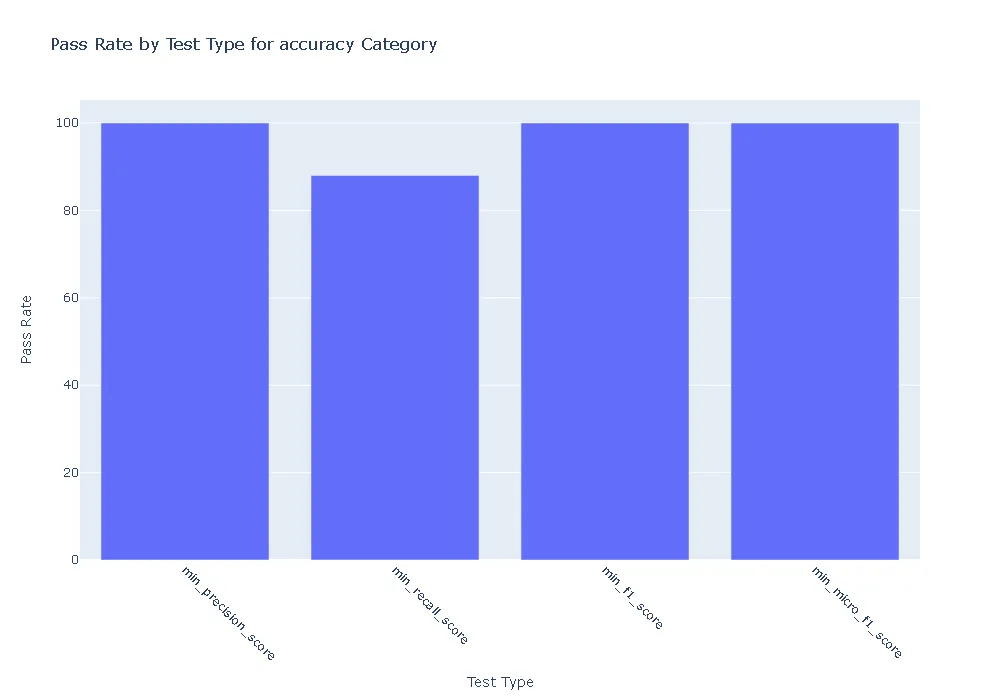
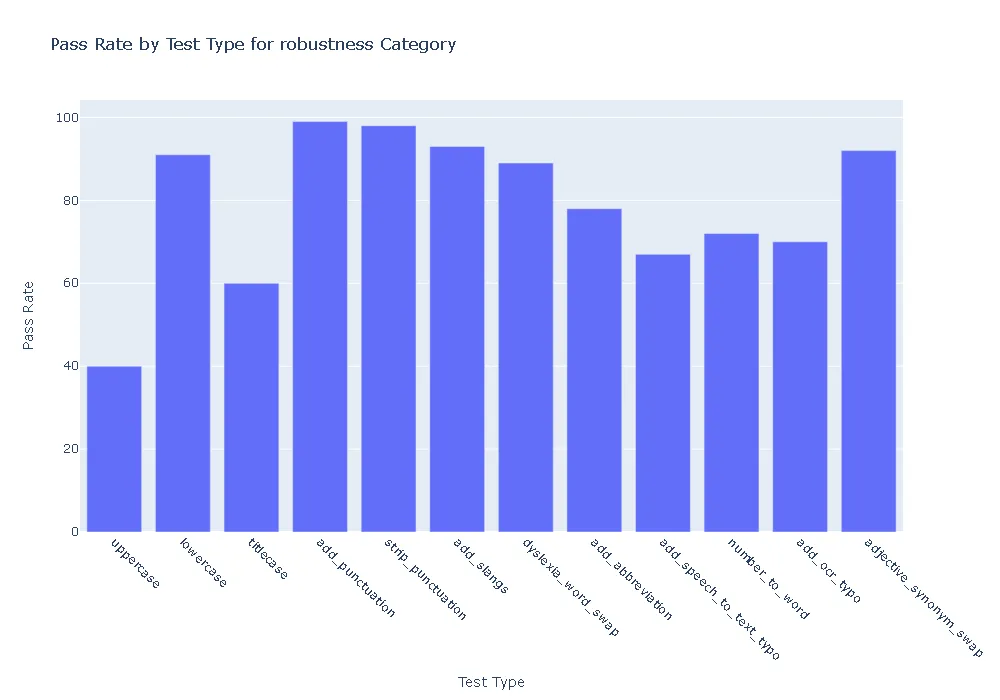
Pass Rate and Fail Count for robustness

Testing med7 model
Instantiate the Harness Class
harness = Harness(
task = "ner",
data={"data_source":"sample-test.conll"},
model={"model":"en_core_med7_lg","hub":"spacy"}
)Test Case Generation and Execution
# Configure the Tests configure_tests() # Call the test configuration function # Generating the test cases harness.generate() # This method automatically generates test cases based on the provided configuration. # Running the tests harness.run() # This method is called after harness.generate() and is used to run all the tests. It returns a pass/fail flag for each test. # Running the tests harness.report()
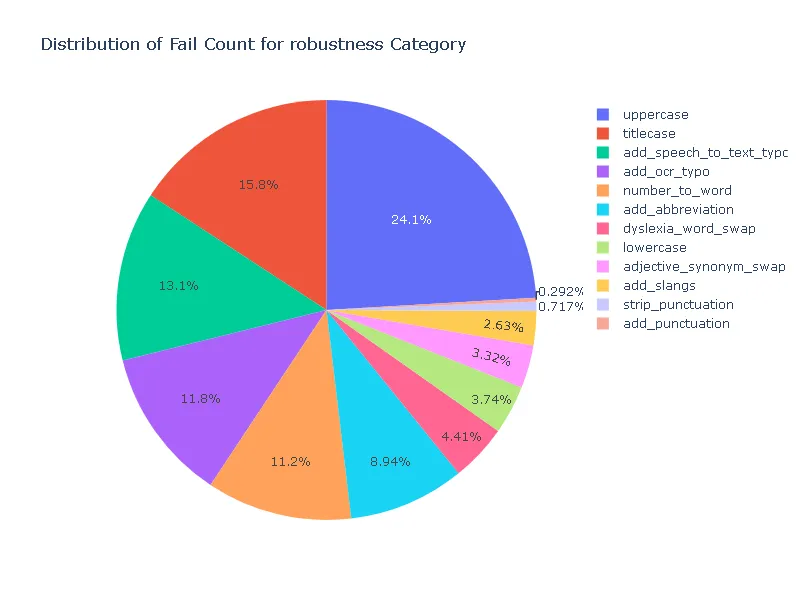
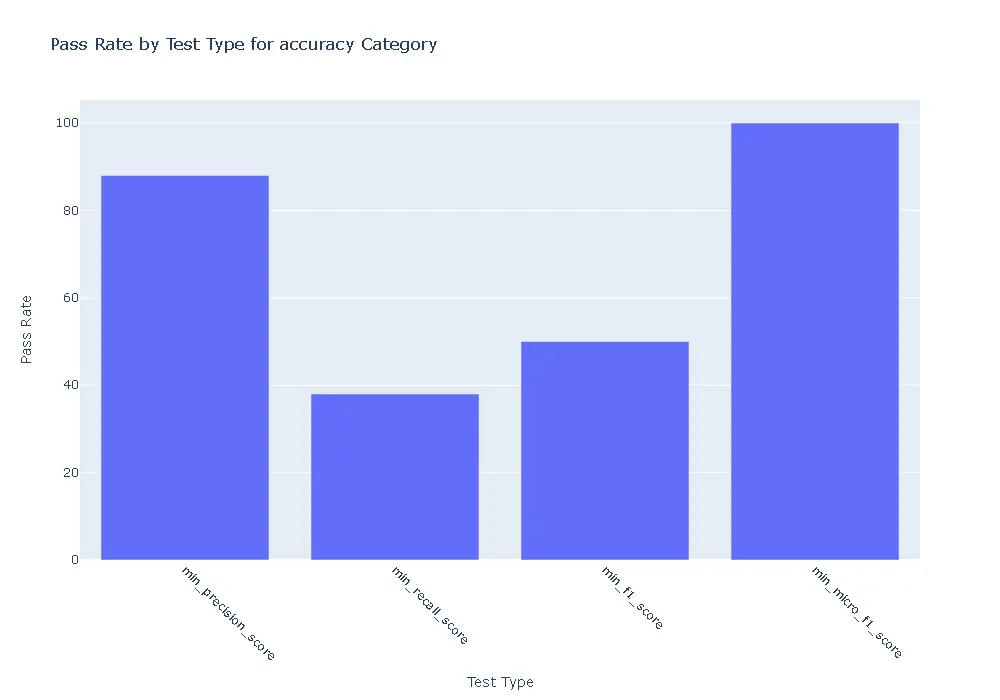
Pass Rate and Fail Count for robustness

Comparing Models
comparing the pass_rate of ner posology from JSL and med7 from spacy.
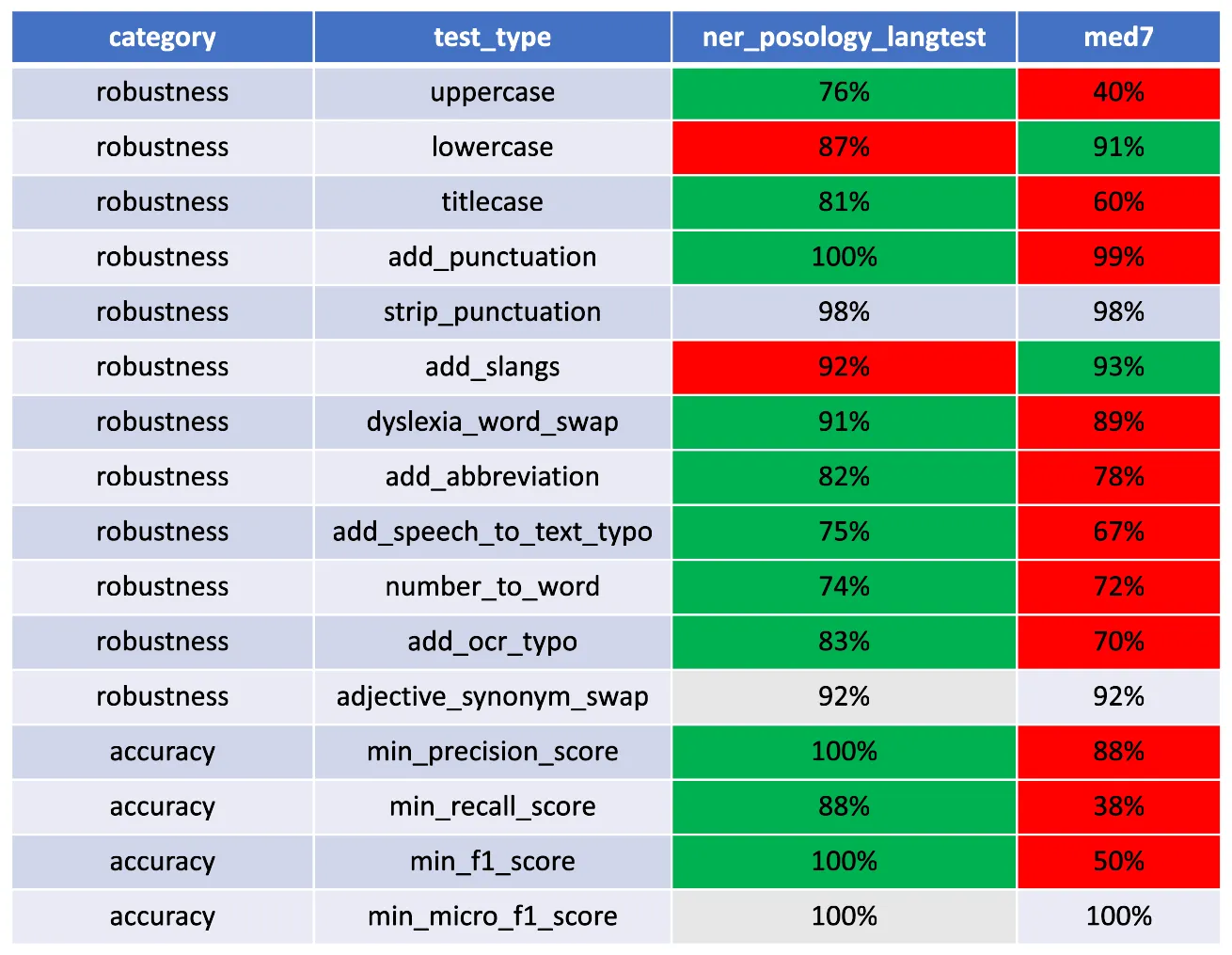
After evaluating the performance of the ner posology and med7 models in terms of Robustness, and Accuracy, here are the findings:
- Accuracy:The ner_posology model demonstrated exceptional accuracy, achieving a perfect pass rate across all accuracy-related tests. In contrast, the med7 model exhibited weaknesses, with failures observed in precision, recall, and F1 score tests.
- Robustness: In terms of robustness, the ner posology model outperformed the med7 model in most test categories. This indicates the ner posology model’s ability to effectively handle a wider range of inputs, highlighting its superior robustness.
Conclusion
In conclusion, while accuracy is undoubtedly crucial, robustness testing takes natural language processing (NLP) models evaluation to the next level by ensuring that models can perform reliably and consistently across a wide array of real-world conditions.
To further enhance the performance of these models, it is crucial to address any identified weaknesses. One potential solution is to consider augmenting the training set using langtest. By incorporating langtest for augmentation, we can potentially improve the models’ generalization and adaptability to diverse patterns and contexts.
To explore the implementation of langtest for augmentation, I recommend referring to the Generating Augmentations section of the langtest website. This approach may lead to more robust models that can deliver even better results in various applications.
